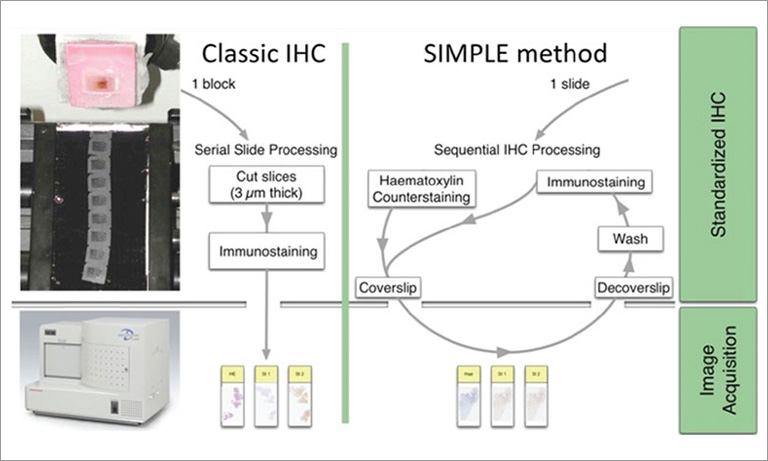
Why NanoZoomer
Products
Solutions
Case study
Resources
United Kingdom (EN)
Select your region or country.
Virtual microscopy system for cervical cytology slides
Cytological screening for cervical pre-cancers has led to a reduction of cervical cancer incidence. Worldwide it is a subjective and variable method with low single-test sensitivity. New biomarkers like p16 that specifically highlight abnormal cervical cells can improve cytology performance. Virtual microscopy offers an ideal platform for assisted evaluation and archiving of biomarkerstained slides. Current automated Pap evaluation systems are based on conventional microscopes and assist the cytology evaluation process by reducing the number of normal slides to be analyzed. Yet these systems are not suited for generating and archiving virtual slides at a large scale. Expert cytologists and slides need to be at the same physical location and multi-observer evaluations require physical sending of slides. A slide scanner on the other hand, e.g. Hamamatsu NanoZoomerⓇ 2.0-HT, is capable of generating digital microscopic images of cytological slides at different focus levels to visualize three-dimensional structures within the cytological sample. After completion of image acquisition, digital images can be studied in seamless levels of magnification. Storage of these digital slides on an image server allows for viewing independent of the physical location of pathologist and sample.
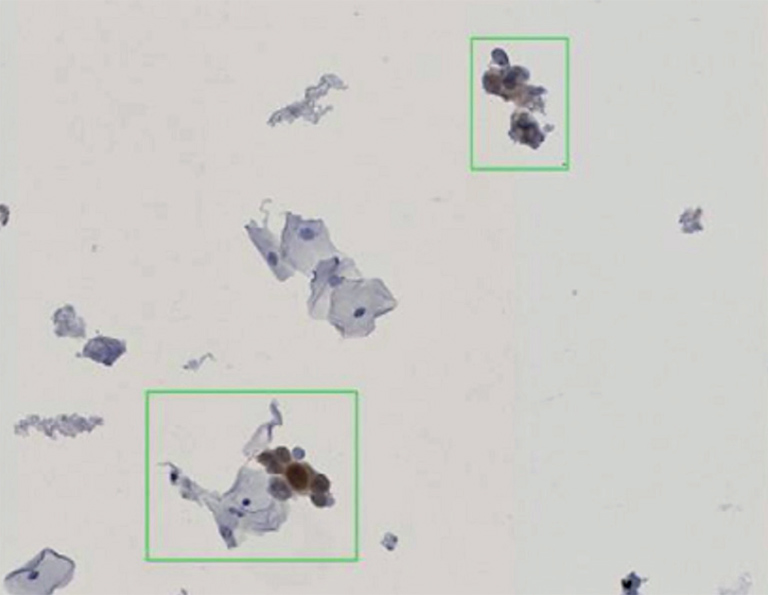
Figure 1: Stained cells, detected by CyTIGA
“Based on a NanoZoomer 2.0-HT we developed the prototype of a system capable of scanning cytological slides, automatically separating cell clusters and individual cells, detecting nuclei and cytoplasm, and identifying biomarkerstained cells”, tells us PD Dr.-Ing. Niels Grabe, Scientific Head of the TIGA center. “We implemented a virtual microscopy system allowing highly efficient automated prescreening and archiving of biomarker-stained slides.”
According to Bernd Lahrmann (PhD student) all slides were scanned with 20× resolution (0.46 µm/pixel) resulted in image sizes of 65K × 60K pixels. For the image database the NDPⓇ Serve image server of Hamamatsu Photonics was used. “We developed image processing algorithms capable to detect positive stained cells on the slides, like the one shown in Fig.1” he explains his work. “After removal of artifacts, based on the specific HSV channel values determined earlier, the algorithm was tested with reference samples and yielded very good agreement with the known manual scoring results. To make it more user-friendly, we developed a novel web server application (CyTIGA server) which provides a user interface for cytological evaluation as shown in Fig.2, Bernd Lahrmann explains.“

Figure 2: Scoring results of the CyTIGA server
“The CyTIGA server provides one-click diagnostic decision-buttons but also offers virtual slide browsing features. This allows interactive navigation through the slide guided by a pre-computed navigation route visiting all or only biomarker-stained objects. The one-click decision-buttons allow fast and simple scoring of each object by an expert.”
But of course such an automatic evaluation algorithm is only useful for daily diagnosis, if it excels manual scoring, which is always subjective and prone to errors by cells not detected or the fact, that not the whole slide is used for evaluation. First tests were done with positive results in favor of the CyTIGA system. The sensitivity of the algorithm to detect biomarker-stained cells was very high (89.1–100%) on the full slides and even higher than the manual evaluation (84.4–98.2%) with specificity above 98.9% for both, manual and automatic analysis.). Furthermore, CyTIGA stores statistical information like number of cells, number of positive events etc., information not available for further analysis after manual scoring.
The TIGA Center is a cooperative project which started in 2007, at the University Heidelberg, with the goal of establishing a bioinformatics platform dedicated to the quantitative analysis and modeling of tissues. A strong emphasis is placed on clinically relevant research projects. At the heart of the TIGA’s technology platform are automated microscopic scanners for whole slide imaging of glass slides. By integrating such imaging systems in a technical pipeline ranging from organotypic in vitro cell cultures to computational tissue modeling, the TIGA generates a wealth of yet unexploited clinically highly relevant tissue data.
PD Dr.-Ing. Niels Grabe is the scientific head of the TIGA center, Dipl.-Bioinf. Bernd Lahrmann is PhD student in medical informatics at the TIGA center.
For further information see “A virtual microscopy system to scan, evaluate and archive biomarker enhanced cervical cytology slides”, Cellular Oncology 32 (2010) 109-119 and http://tigacenter.bioquant.uni-heidelberg.de


Other research case study
- Confirmation
-
It looks like you're in the . If this is not your location, please select the correct region or country below.
You're headed to Hamamatsu Photonics website for GB (English). If you want to view an other country's site, the optimized information will be provided by selecting options below.
In order to use this website comfortably, we use cookies. For cookie details please see our cookie policy.
- Cookie Policy
-
This website or its third-party tools use cookies, which are necessary to its functioning and required to achieve the purposes illustrated in this cookie policy. By closing the cookie warning banner, scrolling the page, clicking a link or continuing to browse otherwise, you agree to the use of cookies.
Hamamatsu uses cookies in order to enhance your experience on our website and ensure that our website functions.
You can visit this page at any time to learn more about cookies, get the most up to date information on how we use cookies and manage your cookie settings. We will not use cookies for any purpose other than the ones stated, but please note that we reserve the right to update our cookies.
1. What are cookies?
For modern websites to work according to visitor’s expectations, they need to collect certain basic information about visitors. To do this, a site will create small text files which are placed on visitor’s devices (computer or mobile) - these files are known as cookies when you access a website. Cookies are used in order to make websites function and work efficiently. Cookies are uniquely assigned to each visitor and can only be read by a web server in the domain that issued the cookie to the visitor. Cookies cannot be used to run programs or deliver viruses to a visitor’s device.
Cookies do various jobs which make the visitor’s experience of the internet much smoother and more interactive. For instance, cookies are used to remember the visitor’s preferences on sites they visit often, to remember language preference and to help navigate between pages more efficiently. Much, though not all, of the data collected is anonymous, though some of it is designed to detect browsing patterns and approximate geographical location to improve the visitor experience.
Certain type of cookies may require the data subject’s consent before storing them on the computer.
2. What are the different types of cookies?
This website uses two types of cookies:
- First party cookies. For our website, the first party cookies are controlled and maintained by Hamamatsu. No other parties have access to these cookies.
- Third party cookies. These cookies are implemented by organizations outside Hamamatsu. We do not have access to the data in these cookies, but we use these cookies to improve the overall website experience.
3. How do we use cookies?
This website uses cookies for following purposes:
- Certain cookies are necessary for our website to function. These are strictly necessary cookies and are required to enable website access, support navigation or provide relevant content. These cookies direct you to the correct region or country, and support security and ecommerce. Strictly necessary cookies also enforce your privacy preferences. Without these strictly necessary cookies, much of our website will not function.
- Analytics cookies are used to track website usage. This data enables us to improve our website usability, performance and website administration. In our analytics cookies, we do not store any personal identifying information.
- Functionality cookies. These are used to recognize you when you return to our website. This enables us to personalize our content for you, greet you by name and remember your preferences (for example, your choice of language or region).
- These cookies record your visit to our website, the pages you have visited and the links you have followed. We will use this information to make our website and the advertising displayed on it more relevant to your interests. We may also share this information with third parties for this purpose.
Cookies help us help you. Through the use of cookies, we learn what is important to our visitors and we develop and enhance website content and functionality to support your experience. Much of our website can be accessed if cookies are disabled, however certain website functions may not work. And, we believe your current and future visits will be enhanced if cookies are enabled.
4. Which cookies do we use?
There are two ways to manage cookie preferences.
- You can set your cookie preferences on your device or in your browser.
- You can set your cookie preferences at the website level.
If you don’t want to receive cookies, you can modify your browser so that it notifies you when cookies are sent to it or you can refuse cookies altogether. You can also delete cookies that have already been set.
If you wish to restrict or block web browser cookies which are set on your device then you can do this through your browser settings; the Help function within your browser should tell you how. Alternatively, you may wish to visit www.aboutcookies.org, which contains comprehensive information on how to do this on a wide variety of desktop browsers.
5. What are Internet tags and how do we use them with cookies?
Occasionally, we may use internet tags (also known as action tags, single-pixel GIFs, clear GIFs, invisible GIFs and 1-by-1 GIFs) at this site and may deploy these tags/cookies through a third-party advertising partner or a web analytical service partner which may be located and store the respective information (including your IP-address) in a foreign country. These tags/cookies are placed on both online advertisements that bring users to this site and on different pages of this site. We use this technology to measure the visitors' responses to our sites and the effectiveness of our advertising campaigns (including how many times a page is opened and which information is consulted) as well as to evaluate your use of this website. The third-party partner or the web analytical service partner may be able to collect data about visitors to our and other sites because of these internet tags/cookies, may compose reports regarding the website’s activity for us and may provide further services which are related to the use of the website and the internet. They may provide such information to other parties if there is a legal requirement that they do so, or if they hire the other parties to process information on their behalf.
If you would like more information about web tags and cookies associated with on-line advertising or to opt-out of third-party collection of this information, please visit the Network Advertising Initiative website http://www.networkadvertising.org.
6. Analytics and Advertisement Cookies
We use third-party cookies (such as Google Analytics) to track visitors on our website, to get reports about how visitors use the website and to inform, optimize and serve ads based on someone's past visits to our website.
You may opt-out of Google Analytics cookies by the websites provided by Google:
https://tools.google.com/dlpage/gaoptout?hl=en
As provided in this Privacy Policy (Article 5), you can learn more about opt-out cookies by the website provided by Network Advertising Initiative:
http://www.networkadvertising.org
We inform you that in such case you will not be able to wholly use all functions of our website.
Close