Why NanoZoomer
Products
Solutions
Case study
Resources
United Kingdom (EN)
Select your region or country.
FISHing of gut microbiota – microbiology in health and diseases
BIOASTER, a new model for technology innovation in microbiology
The BIOASTER Technology Research Institute (TRI) was created in April 2012 by the Institut Pasteur and Lyonbiopole health competitiveness cluster, following the initiative of the French Government through the “Investissements d’Avenir” Program.
BIOASTER is leading collaborative projects that bring together academics, startups, SMEs and industrial groups, and is developing a unique technological and innovative model in order to overcome technological bottlenecks and explore new avenues applied to microbiology in health and diseases. In this context, the Preclinical Models & Imaging Unit tested the Hamamatsu NanoZoomerⓇ S60 Digital slide scanner with its fluorescence module, to evaluate its relevance for bacteria imaging in relation to the study of host-microbiota and microbes-microbes interactions.
Bringing the gut microbiota into focus
Histological processing can alter tissues so several sections are often placed on the same microscopy slide in order to maximize the acquisition of views. One great advantage of the NanoZoomer is its capacity to scan the whole microscopy slide with the possibility afterwards to zoom anywhere within the scanned image to look just as well at a whole tissue section (Figure 1) as at a specific subcellular site (Figure 3).
In addition to high quality magnifi cation, the NanoZoomer offers two essential features, an automated focus with the possibility to delete or add manually focus check points on the epithelium, and Z-stacking for samples with 3D structures such as bacteria. We found that 0.5 µm z-steps travelling from -0.5 µm (below focus point) to +1.5 µm (above focus point) were optimal for mouse colon sections of 4 µm.
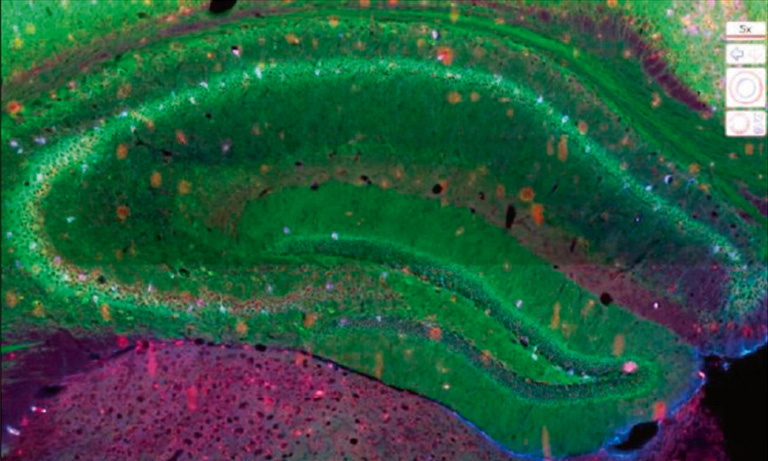
Another great value of the NanoZoomer is the possibility to detect 6 different fluorescent probes or more, optimize individual intensity and potentially modify colors to improve clarity, allowing for co-staining of different targets, but it also makes it possible to overcome some limitations due to the green autofluorescence of the intestinal epithelium (1) and lumen, which might mask bacteria specific signals (Figure 1).
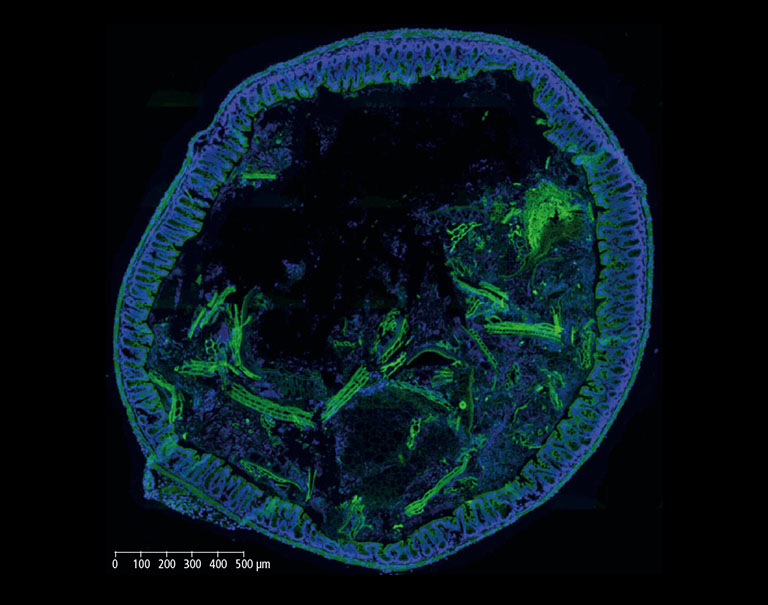
Figure 1: Whole mouse colon section stained with DAPI (blue) showing high level of green autofluorescence from the gut epithelium and lumen, which can be a limitation for imaging samples with green channel (FITC). 20x objective, 4x view.
Imaging of Lactobacilli detected by specific fluorescent in situ hybridization (FISH) in mouse colon sections
Increased evidence that gut microbiota is a key factor in human health and diseases opens a new golden age in microbiology. From pathologies to phenotypical features, its range of action holds promises which have to be unveiled in the near future. However, the gut microbiota is an ecosystem that has proved itself to be a lot more complicated than what was expected. As a result, the development of simplified models, but conserving the key features of original models, is of high necessity in enabling scientists to study the causal link between microbiota changes and the host response (2, 3).
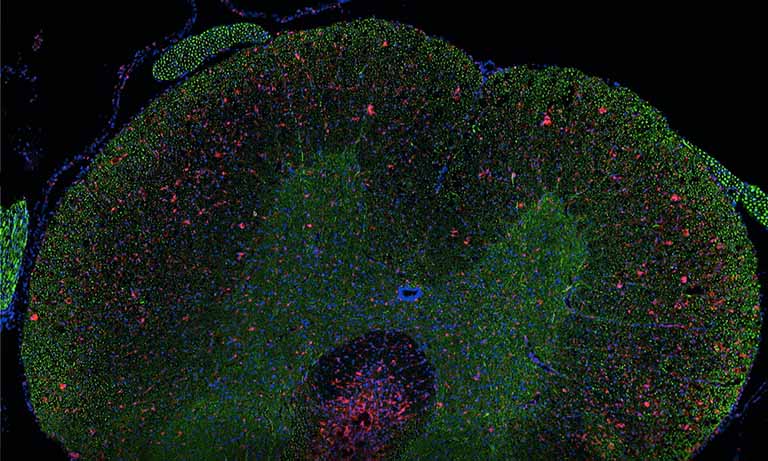
In the context of an internal innovation project at BIOASTER which aimed to establish such a gnotobiotic mouse model, we set up FISH imaging activities in order to highlight host-microbiota and microbes-microbes interactions. Hereinafter, we show images of a mouse colon section stained with the FISH probe LGC0355 targeting Lactobacilli, Firmicutes (Figure 2), which are among the most common bacteria in the mouse gut, but also in the intestine of humans and widely used as probiotics.
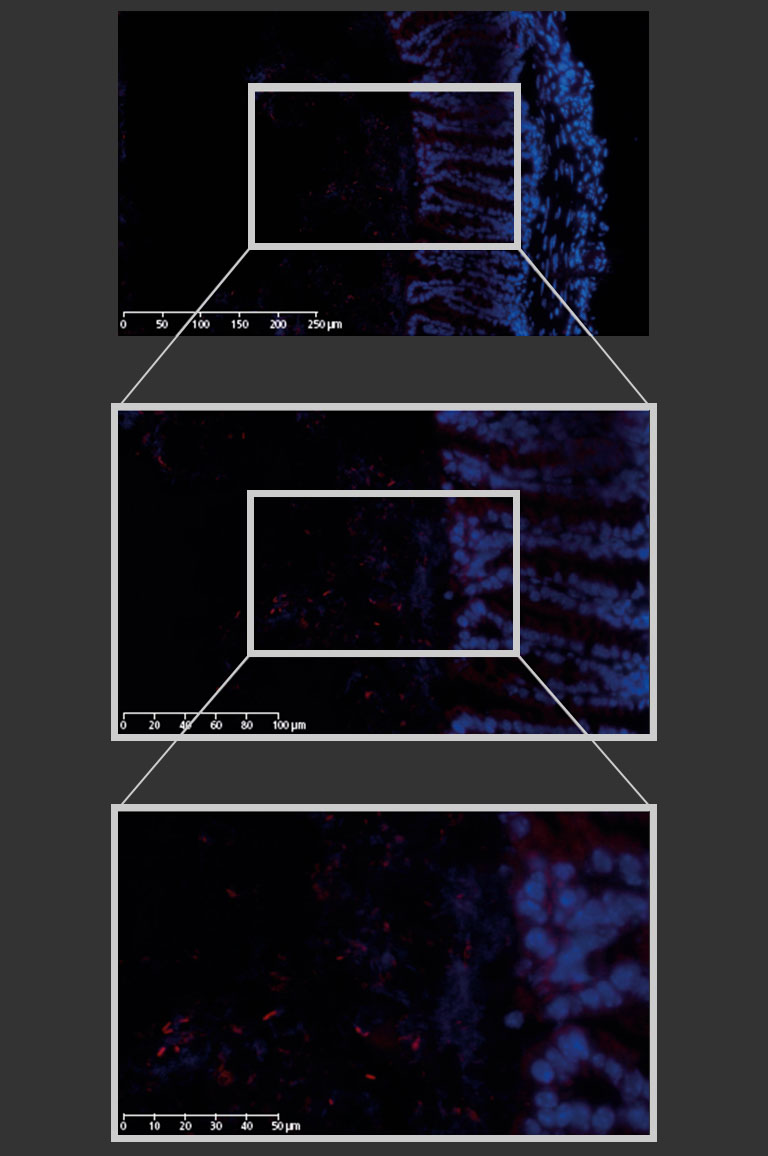
Figure 2: Mouse colon section showing epithelial cell nuclei stained with DAPI (blue) and Lactobacilli distribution within the lumen, detected by the FISH probe LGC0355 labeled with the fluorophore CY5 (red). 20x objective (above), magnification to 40x (middle) and additional numerical zoom to 80x (below).
Combining immunofluorescence (IF) and bacterial FISH to highlight the intestinal barrier
Next, we additionally stained the mucosal barrier in order to inspect the border between the intestinal epithelium and the microbes (Figure 3). The mucus secreted by Goblet cells protects the epithelial cells against damage, but also accommodates Lactobacilli which have adhesion properties on mucins, allowing suitable interaction with the host to confer health benefits(4).
In conclusion, the NanoZoomer S60 Digital slide scanner provides high quality fluorescence imaging of gut microbiota comparable to confocal microscopy, with improved reproducibility and time management thanks to automated scanning and a proper slide capacity. On top of that, the same equipment allows brightfield imaging, and easy-to-use software offers full options analysis, including interactive data sharing with collaborators.
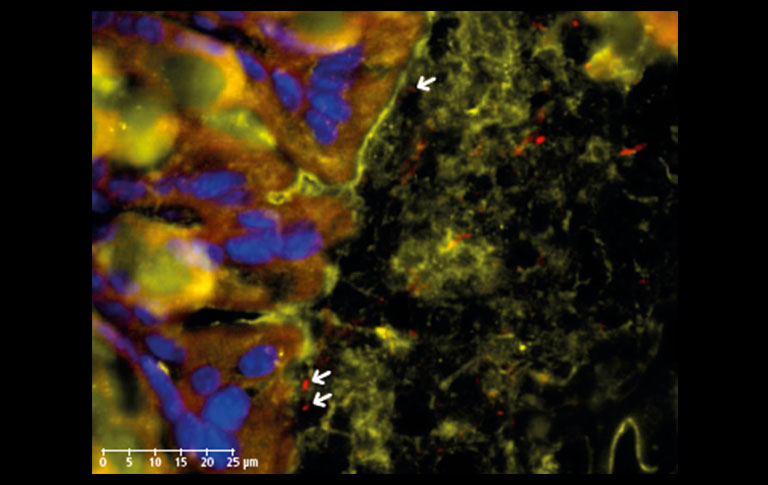
Figure 3: IF staining of Muc2 secreted by Goblet cells (orange, TxRed) in mouse colon section showing the adhesion of Lactobacilli (red, CY5) to host surfaces (epithelial cell nuclei stained with DAPI, blue) through mucus-microbe interactions (arrows). 20x objective, 80x view.
For further information see:
- Monici M. (2005) “Cell and tissue autofl uorescence research and diagnostic applications.” Biotechnol Annu Rev 11, 227.
- Stappenbeck TS, Virgin HW. (2016) “Accounting for reciprocal hostmicrobiome interactions in experimental science.” Nature 534, 191.
- Martin R, Bermudez-Humaran LG, Langella P. (2016) “Gnotobiotic Rodents: An In Vivo Model for the study of Microbe-Microbe Interactions.” Front. Microbiol. 7, 409.
- Van Tassell ML, Miller MJ. (2011) “Lactobacillus Adhesion to Mucus.” Nutrients 3, 613.
FISHing of gut microbiota - microbiology in health and diseases [3.6 MB/PDF]

Other research case study
- Confirmation
-
It looks like you're in the . If this is not your location, please select the correct region or country below.
You're headed to Hamamatsu Photonics website for GB (English). If you want to view an other country's site, the optimized information will be provided by selecting options below.
In order to use this website comfortably, we use cookies. For cookie details please see our cookie policy.
- Cookie Policy
-
This website or its third-party tools use cookies, which are necessary to its functioning and required to achieve the purposes illustrated in this cookie policy. By closing the cookie warning banner, scrolling the page, clicking a link or continuing to browse otherwise, you agree to the use of cookies.
Hamamatsu uses cookies in order to enhance your experience on our website and ensure that our website functions.
You can visit this page at any time to learn more about cookies, get the most up to date information on how we use cookies and manage your cookie settings. We will not use cookies for any purpose other than the ones stated, but please note that we reserve the right to update our cookies.
1. What are cookies?
For modern websites to work according to visitor’s expectations, they need to collect certain basic information about visitors. To do this, a site will create small text files which are placed on visitor’s devices (computer or mobile) - these files are known as cookies when you access a website. Cookies are used in order to make websites function and work efficiently. Cookies are uniquely assigned to each visitor and can only be read by a web server in the domain that issued the cookie to the visitor. Cookies cannot be used to run programs or deliver viruses to a visitor’s device.
Cookies do various jobs which make the visitor’s experience of the internet much smoother and more interactive. For instance, cookies are used to remember the visitor’s preferences on sites they visit often, to remember language preference and to help navigate between pages more efficiently. Much, though not all, of the data collected is anonymous, though some of it is designed to detect browsing patterns and approximate geographical location to improve the visitor experience.
Certain type of cookies may require the data subject’s consent before storing them on the computer.
2. What are the different types of cookies?
This website uses two types of cookies:
- First party cookies. For our website, the first party cookies are controlled and maintained by Hamamatsu. No other parties have access to these cookies.
- Third party cookies. These cookies are implemented by organizations outside Hamamatsu. We do not have access to the data in these cookies, but we use these cookies to improve the overall website experience.
3. How do we use cookies?
This website uses cookies for following purposes:
- Certain cookies are necessary for our website to function. These are strictly necessary cookies and are required to enable website access, support navigation or provide relevant content. These cookies direct you to the correct region or country, and support security and ecommerce. Strictly necessary cookies also enforce your privacy preferences. Without these strictly necessary cookies, much of our website will not function.
- Analytics cookies are used to track website usage. This data enables us to improve our website usability, performance and website administration. In our analytics cookies, we do not store any personal identifying information.
- Functionality cookies. These are used to recognize you when you return to our website. This enables us to personalize our content for you, greet you by name and remember your preferences (for example, your choice of language or region).
- These cookies record your visit to our website, the pages you have visited and the links you have followed. We will use this information to make our website and the advertising displayed on it more relevant to your interests. We may also share this information with third parties for this purpose.
Cookies help us help you. Through the use of cookies, we learn what is important to our visitors and we develop and enhance website content and functionality to support your experience. Much of our website can be accessed if cookies are disabled, however certain website functions may not work. And, we believe your current and future visits will be enhanced if cookies are enabled.
4. Which cookies do we use?
There are two ways to manage cookie preferences.
- You can set your cookie preferences on your device or in your browser.
- You can set your cookie preferences at the website level.
If you don’t want to receive cookies, you can modify your browser so that it notifies you when cookies are sent to it or you can refuse cookies altogether. You can also delete cookies that have already been set.
If you wish to restrict or block web browser cookies which are set on your device then you can do this through your browser settings; the Help function within your browser should tell you how. Alternatively, you may wish to visit www.aboutcookies.org, which contains comprehensive information on how to do this on a wide variety of desktop browsers.
5. What are Internet tags and how do we use them with cookies?
Occasionally, we may use internet tags (also known as action tags, single-pixel GIFs, clear GIFs, invisible GIFs and 1-by-1 GIFs) at this site and may deploy these tags/cookies through a third-party advertising partner or a web analytical service partner which may be located and store the respective information (including your IP-address) in a foreign country. These tags/cookies are placed on both online advertisements that bring users to this site and on different pages of this site. We use this technology to measure the visitors' responses to our sites and the effectiveness of our advertising campaigns (including how many times a page is opened and which information is consulted) as well as to evaluate your use of this website. The third-party partner or the web analytical service partner may be able to collect data about visitors to our and other sites because of these internet tags/cookies, may compose reports regarding the website’s activity for us and may provide further services which are related to the use of the website and the internet. They may provide such information to other parties if there is a legal requirement that they do so, or if they hire the other parties to process information on their behalf.
If you would like more information about web tags and cookies associated with on-line advertising or to opt-out of third-party collection of this information, please visit the Network Advertising Initiative website http://www.networkadvertising.org.
6. Analytics and Advertisement Cookies
We use third-party cookies (such as Google Analytics) to track visitors on our website, to get reports about how visitors use the website and to inform, optimize and serve ads based on someone's past visits to our website.
You may opt-out of Google Analytics cookies by the websites provided by Google:
https://tools.google.com/dlpage/gaoptout?hl=en
As provided in this Privacy Policy (Article 5), you can learn more about opt-out cookies by the website provided by Network Advertising Initiative:
http://www.networkadvertising.org
We inform you that in such case you will not be able to wholly use all functions of our website.
Close