Why NanoZoomer
Products
Solutions
Case study
Resources
United States (EN)
Select your region or country.
Quantifying virtual co-localization of tissue-based biomarkers
DIAPath, Center for Microscopy and Molecular imaging (www.cmmi.be, Gosselies, Belgium)
Whole slide imaging (WSI) using a NanoZoomerⓇ 2.0-HT is an essential step of the integrated solution which is developed since 2010 by DIAPath, the digital pathology platform of the CMMI (Gosselies, Belgium), ensuring the analysis and the validation of tissue-based biomarkers. This solution combines immunohistochemistry (IHC), special staining, WSI, image processing and data analysis. It is based on standardized laboratory procedures and quality controls ensuring reproducibility and traceability.
Extracting relevant information from actors involved in complex biological processes (such as cancers or treatment responses) requires targeting different antigens simultaneously. Multichromogenic (brightfield) IHC is used for multiple antigen labeling on the same slide but suffers from limitations. In addition to antibody cross-reaction problems, this approach also prevents the analysis of proteins expressed in the same cellular compartment (because of color merging or masking). We thus developed an alternative which consists in applying standard IHC on consecutive (also labeled “serial”) tissues sections to target different proteins (see Fig. 1, left-hand panel). These slides were then digitalized and superimposed by a registration algorithm. Our approach uses the fact that histological structures (such as glands or epithelium) are often well conserved across a few serial slides because the slide thickness obtained with microtomy is 3 to 5 μm (i.e. smaller than the mean size of human cells).
To superimpose the serial slide images we developed an efficient 2-step registration algorithm [1]. The first, low-resolution, registration step is applied on the 1X equivalent magnification images (showing the whole slides to register in small-sized images such as 4,000 by 3,000 pixels). Briefly, this step enables to roughly superimpose two virtual slides on the basis of the slice outlines, holes and/or large histological structures. After this first step a series of high-resolution registrations were independently performed on the set of high-resolution (i.e. 20X equivalent) fields of view constituting a whole slide image.
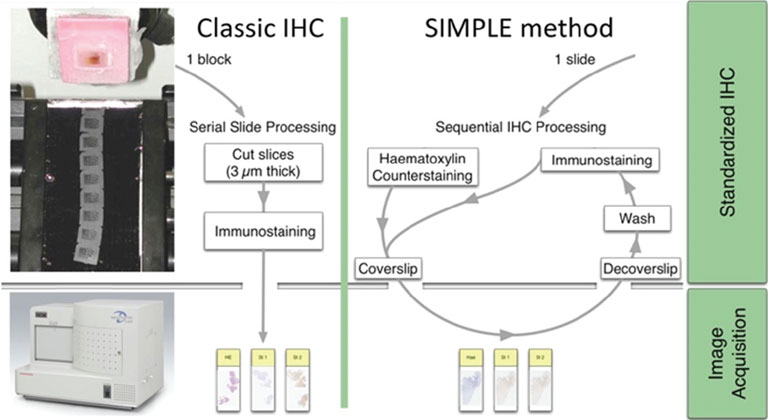
Figure 1: IHC on serial tissue slides (left-hand panel) and SIMPLE technique applied on the same slide (right-hand panel). Both provide slide images to superimpose for colocalizing different antigen expressions.
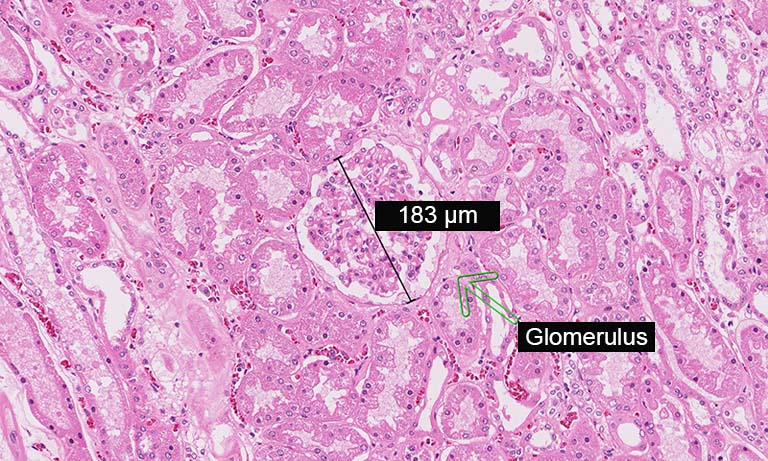
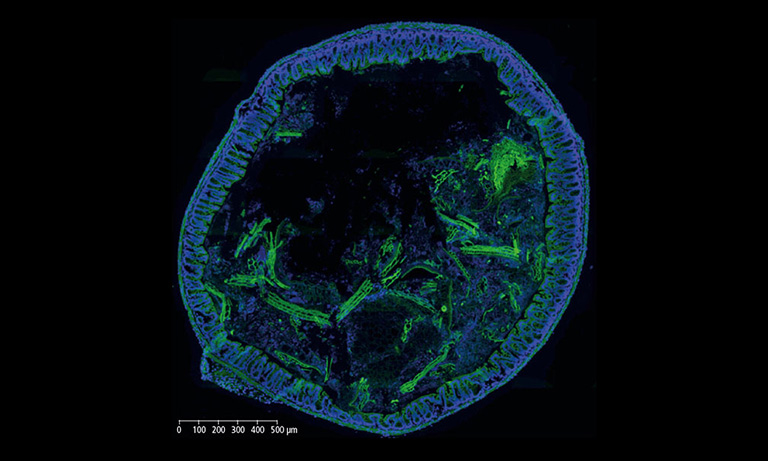
We evaluated image registration accuracy by measuring the distances between pairs of control points (located on each image) which should match if the images were perfectly superimposed. Our approach shows accuracy levels compatible with biomarker colocalization characterization. Indeed, on serial slides the distances between control points were evaluated to be at most 20 μm in presence of histological structures in the tissue (e.g. in colonic tissue, see Fig. 2). We then implemented a method to extract biomarker colocalization measurements taking the level of registration accuracy into account and validated our complete procedure by comparison to colocalization information obtained by means of double staining (with different cell locations, Fig. 2).
Because registration accuracy was lower for tissue without histological structures (e.g. the distances between control points being at most 80 μm for serial brain tumor slides), we developed a sequential IHC technique applied on the same slide (Fig. 1, right-hand panel). This “Sequential Immunoperoxidase Labeling and Erasing” (SIMPLE) method is based on cycles of staining/digitization/erasing, where after IHC staining and slide digitization, staining is “washed” through an antibody elution technique. We improved the original SIMPLE method by including a new elution methodology which preserves tissue and antigen epitopes for the next staining step. We successfully used this approach to identify antigens expressed in the same cellular compartment of high-grade glioma samples. We tested our registration method on the virtual slides so obtained and achieved very good results, i.e. about 5 μm of distance between control points [1].
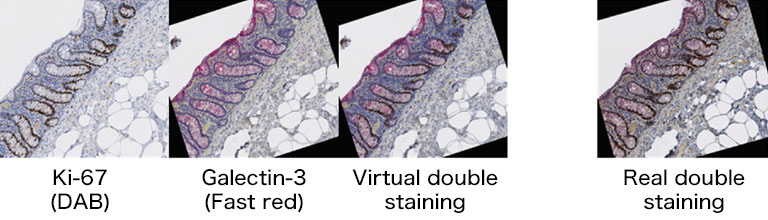
Figure 2: Virtual double IHC staining: Result of image registration applied to serial slides independently submitted to IHC and compared to double IHC staining (targeting Ki-67 and galectin-3) on a colonic tissue sample.
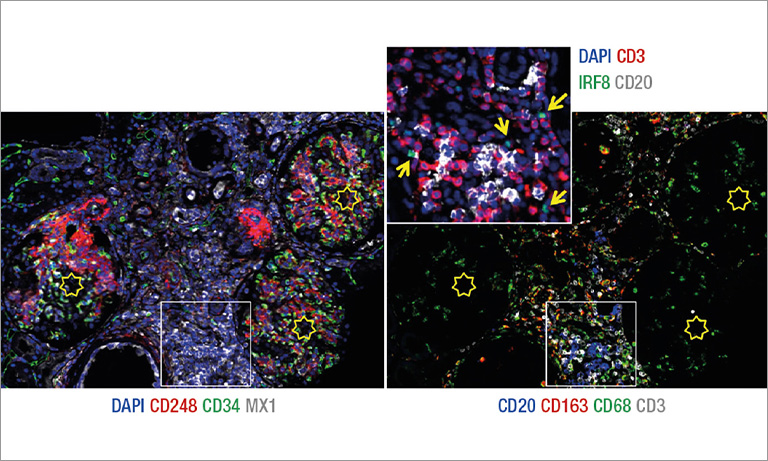
Afterwards we adapted our registration process to tissue microarray (TMA) slides for high-throughput analysis of biomarker colocalization. Registration is easier for these slide images because TMA assembles cores extracted from tissue blocks in array fashion. As expected, we obtained very accurate registration results on serial TMA slides, even for unstructured tissue (i.e. distance between control points less than 10 μm) [2]. Fig. 3 shows the different steps of the process requiring (1) the correct identification of the circular samples (from the cores) on each TMA image, (2) the registration of core images extracted from serial slide images and (3) the computation of a colocalization map on small squared tiles from which a global colocalization measurement (adapted from immunofluorescence) can be computed [1,2]. A first (not illustrated) step consists in computing in each tile the value of a staining feature characterizing each IHC marker, e.g. the labeling index (LI), which is the percentage of positive (i.e. DAB-stained) tissue area [3]. The colocalization map evidences local staining overlap, which increases when both LI increases in a tile (see Fig. 3.3). The value of the global overlap index (r0) is the sum of the tile contributions and takes values between 0 and 1.
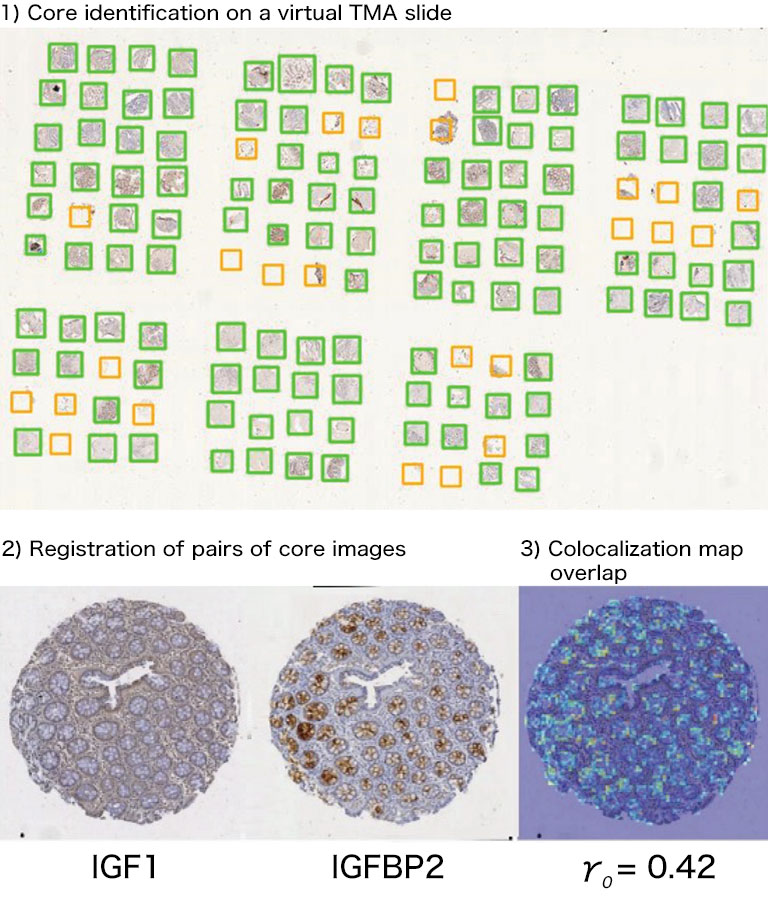
Figure 3: IHC staining colocalization on serial TMA slides showing IGF-I and IGFBP2 IHC expression in colonic tissue. After 1) core identification on each TMA image, 2) core images are extracted and registered to compute 3) staining overlap map and index (r0). In the map blue indicates no overlap whereas yellow to red show increasing overlap levels.
References:
[1] X. Moles Lopez, et al. Registration of whole immunohistochemical slide images: an efficient way to characterize biomarker colocalization. J Am Med Inform Assoc 22(1):86–99, 2015.
[2] Y.-R. Van Eycke, et al. High-throughput analysis of tissue-based biomarkers in digital pathology. In: 2015 37th Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. IEEE, Aug. 2015, pp. 7732–7735.
[3] C. Decaestecker, et al. Requirements for the valid quantification of immunostains on tissue microarray materials using image analysis. Proteomics 9(19):4478–94, 2009.
Yves-Rémi Van Eycke (1,2), Justine Allard (1), Cédric Balsat (1), Isabelle Salmon (1,3), Christine Decaestecker (1,2)
(1) DIAPath, Center for Microscopy and Molecular Imaging, Université Libre de Bruxelles (ULB), Gosselies, Belgium
(2) Laboratories of Image, Signal processing & Acoustics, ULB, Brussels, Belgium
(3) Department of Pathology, Erasme Hospital, ULB, Brussels, Belgium
For further information, please contact: amarcowycz@hamamatsu.be
Quantifying virtual co-localization of tissue-based biomarkers [11.5 MB/PDF]


Other research case study
- Confirmation
-
It looks like you're in the . If this is not your location, please select the correct region or country below.
You're headed to Hamamatsu Photonics website for US (English). If you want to view an other country's site, the optimized information will be provided by selecting options below.
In order to use this website comfortably, we use cookies. For cookie details please see our cookie policy.
- Cookie Policy
-
This website or its third-party tools use cookies, which are necessary to its functioning and required to achieve the purposes illustrated in this cookie policy. By closing the cookie warning banner, scrolling the page, clicking a link or continuing to browse otherwise, you agree to the use of cookies.
Hamamatsu uses cookies in order to enhance your experience on our website and ensure that our website functions.
You can visit this page at any time to learn more about cookies, get the most up to date information on how we use cookies and manage your cookie settings. We will not use cookies for any purpose other than the ones stated, but please note that we reserve the right to update our cookies.
1. What are cookies?
For modern websites to work according to visitor’s expectations, they need to collect certain basic information about visitors. To do this, a site will create small text files which are placed on visitor’s devices (computer or mobile) - these files are known as cookies when you access a website. Cookies are used in order to make websites function and work efficiently. Cookies are uniquely assigned to each visitor and can only be read by a web server in the domain that issued the cookie to the visitor. Cookies cannot be used to run programs or deliver viruses to a visitor’s device.
Cookies do various jobs which make the visitor’s experience of the internet much smoother and more interactive. For instance, cookies are used to remember the visitor’s preferences on sites they visit often, to remember language preference and to help navigate between pages more efficiently. Much, though not all, of the data collected is anonymous, though some of it is designed to detect browsing patterns and approximate geographical location to improve the visitor experience.
Certain type of cookies may require the data subject’s consent before storing them on the computer.
2. What are the different types of cookies?
This website uses two types of cookies:
- First party cookies. For our website, the first party cookies are controlled and maintained by Hamamatsu. No other parties have access to these cookies.
- Third party cookies. These cookies are implemented by organizations outside Hamamatsu. We do not have access to the data in these cookies, but we use these cookies to improve the overall website experience.
3. How do we use cookies?
This website uses cookies for following purposes:
- Certain cookies are necessary for our website to function. These are strictly necessary cookies and are required to enable website access, support navigation or provide relevant content. These cookies direct you to the correct region or country, and support security and ecommerce. Strictly necessary cookies also enforce your privacy preferences. Without these strictly necessary cookies, much of our website will not function.
- Analytics cookies are used to track website usage. This data enables us to improve our website usability, performance and website administration. In our analytics cookies, we do not store any personal identifying information.
- Functionality cookies. These are used to recognize you when you return to our website. This enables us to personalize our content for you, greet you by name and remember your preferences (for example, your choice of language or region).
- These cookies record your visit to our website, the pages you have visited and the links you have followed. We will use this information to make our website and the advertising displayed on it more relevant to your interests. We may also share this information with third parties for this purpose.
Cookies help us help you. Through the use of cookies, we learn what is important to our visitors and we develop and enhance website content and functionality to support your experience. Much of our website can be accessed if cookies are disabled, however certain website functions may not work. And, we believe your current and future visits will be enhanced if cookies are enabled.
4. Which cookies do we use?
There are two ways to manage cookie preferences.
- You can set your cookie preferences on your device or in your browser.
- You can set your cookie preferences at the website level.
If you don’t want to receive cookies, you can modify your browser so that it notifies you when cookies are sent to it or you can refuse cookies altogether. You can also delete cookies that have already been set.
If you wish to restrict or block web browser cookies which are set on your device then you can do this through your browser settings; the Help function within your browser should tell you how. Alternatively, you may wish to visit www.aboutcookies.org, which contains comprehensive information on how to do this on a wide variety of desktop browsers.
5. What are Internet tags and how do we use them with cookies?
Occasionally, we may use internet tags (also known as action tags, single-pixel GIFs, clear GIFs, invisible GIFs and 1-by-1 GIFs) at this site and may deploy these tags/cookies through a third-party advertising partner or a web analytical service partner which may be located and store the respective information (including your IP-address) in a foreign country. These tags/cookies are placed on both online advertisements that bring users to this site and on different pages of this site. We use this technology to measure the visitors' responses to our sites and the effectiveness of our advertising campaigns (including how many times a page is opened and which information is consulted) as well as to evaluate your use of this website. The third-party partner or the web analytical service partner may be able to collect data about visitors to our and other sites because of these internet tags/cookies, may compose reports regarding the website’s activity for us and may provide further services which are related to the use of the website and the internet. They may provide such information to other parties if there is a legal requirement that they do so, or if they hire the other parties to process information on their behalf.
If you would like more information about web tags and cookies associated with on-line advertising or to opt-out of third-party collection of this information, please visit the Network Advertising Initiative website http://www.networkadvertising.org.
6. Analytics and Advertisement Cookies
We use third-party cookies (such as Google Analytics) to track visitors on our website, to get reports about how visitors use the website and to inform, optimize and serve ads based on someone's past visits to our website.
You may opt-out of Google Analytics cookies by the websites provided by Google:
https://tools.google.com/dlpage/gaoptout?hl=en
As provided in this Privacy Policy (Article 5), you can learn more about opt-out cookies by the website provided by Network Advertising Initiative:
http://www.networkadvertising.org
We inform you that in such case you will not be able to wholly use all functions of our website.
Close